Abstract
The body protein pool constantly turns over while achieving a dynamic steady state in healthy living organisms to accomplish homeostasis, indicating a close match between the rate of protein synthesis and breakdown (i.e., protein turnover). However, dysregulation of protein turnover in muscle over time leads to muscle wasting conditions such as sarcopenia, cachexia, and heart failure. Furthermore, altered muscle metabolism in obesity, insulin resistance, and diabetes. Therefore, skeletal muscle in wasting conditions is an important target for drug development. Since muscle mass is controlled by the balance between the rate of muscle protein synthesis (MPS) and muscle protein breakdown (MPB), a change in muscle mass can occur due to various permutations of the two rates (i.e., MPS and MPB). Elucidation of molecular mechanisms involved in the control of protein turnover provides invaluable data but misses the dynamic (i.e., kinetic) nature of the proteome, information of which stable isotope tracer methodology can provide. Combining stable isotope tracer methodology and molecular biology tools enables an in-depth understanding of protein metabolism in physiological and pathophysiological conditions, consequently facilitating the development of effective therapeutic approaches (optimal nutrition, exercise, and drugs) to treat muscle wasting diseases. In this review, we will discuss the significance of obtaining kinetic information for a better understanding of protein metabolism, and stable isotope tracer methodologies that enable exploration of muscle protein kinetics and direct muscle mass in vivo. These methodologies can be applied to metabolic research regarding pathophysiological conditions in both humans and animals.
-
Keywords: Kinetics; Metabolic flux analysis; Muscular atrophy
INTRODUCTION
The protein pool in the human body was previously believed to be immobile until the innovative work of Schoenheimer et al. [
1] in 1930s, the pioneer of metabolic isotope tracer technology, who first investigated in vivo protein kinetics using
15N labeled amino acids (AAs) and demonstrated the dynamic nature of body proteins (i.e., protein synthesis and protein breakdown). Entitled “The Dynamic State of Body Constituents”, the dynamic states of living systems were eloquently described in his posthumously assembled book in 1946 as “all constituents of living matter are in a steady state of rapid flux” [
2]. Every molecule in the body turns over at various rates to accomplish “dynamic” homeostasis in the body. The protein pool size can change when an imbalance between rates of protein synthesis and breakdown occurs in normal or pathological processes over time. For example, loss of muscle mass can occur if the rate of protein breakdown is greater than protein synthesis, even if an increased rate of protein synthesis exists compared with the normal condition as long as the rate of protein breakdown is accelerated to a greater extent, as observed in catabolic muscle wasting conditions [
3]. This underscores the importance of simultaneous assessment and understanding the rates of protein synthesis and breakdown. Although elucidation of molecular signaling implicated in protein kinetics is invaluable but limited to “snap-shot” information, the stable isotope tracer methodology can provide information on protein dynamics (kinetics), which are often not reflected in static information such as mRNA abundance, signaling, and protein abundance [
4,
5]. Therefore, assessing protein kinetics on both sides of the protein balance equation is important to better understand the dynamic nature of protein metabolism in conjunction with assessing the molecular signaling. In this review, the dynamic nature of protein metabolism, with an emphasis on skeletal muscle protein dynamics, which is important to understand normal and pathological muscle wasting conditions such as sarcopenia and cachexia, is presented. Then, various stable isotope tracer methodologies that enable assessment of in vivo kinetics of protein dynamics at the muscle and whole-body levels are reviewed using representative AA tracer infusion or deuterium oxide (D
2O), each with unique advantages and disadvantages.
MOTION PICTURES DIFFER FROM STATIC: MUSCLE IS IN A CONSTANT STATE OF TURNOVER
The protein pool size or mass in the body is controlled by the balance between protein synthesis and breakdown rates in the different levels of organisms: specific individual proteins (e.g., titin, myosin light chain, creatine kinase) [
6-
9], subcellular organelles (e.g., contractile and mitochondria proteins) [
10,
11], tissue/organ (e.g., muscle, adipose tissue, liver) [
12-
14], or whole systems [
15-
18]. For example, in healthy adults, muscle mass is relatively well maintained due to the direct result of the close relationship between protein synthesis and breakdown rates. In anabolic states such as growing children or body builders, muscle mass increases because the rate of protein synthesis is greater than protein breakdown. Conversely, in catabolic states such as cancer cachexia, heart failure, and sarcopenia, the breakdown of protein is greater than synthesis. Furthermore, achieving similar protein pool sizes (or muscle mass) in states of different turnover is possible but can lead to changes in the quality of the muscle because muscle quality (i.e., muscle strength normalized to mass [
19]) is positively associated with muscle protein turnover rates [
20]. This indicates that rates of protein turnover within a normal physiological range ensures a constant provision of new “functional” proteins (i.e., protein synthesis) to replace old, nonfunctional, or damaged proteins that must be cleared via the process of protein breakdown. Despite the dynamic nature of muscle proteins in vivo, researchers have extensively relied on assessments of static information, termed “statomics”, such as abundance of metabolites, mRNA, and proteins, or the (in)activity of molecular signaling cascades [
5], which can result in erroneous view of actual metabolic states. Significant mismatches between “statomics” and dynamics (i.e., flux) have been observed in human [
4,
21] and animal [
5,
22] studies. In the following sections, basic principles and applications (to research) of stable isotope tracer methodology to assess in vivo metabolic flux rates are reviewed, with a focus on human muscle dynamics, after briefly discussing how this understanding can be used for drug development in muscle wasting conditions.
IMPLICATIONS FOR DEVELOPMENT OF DRUGS TO TREAT MUSCLE WASTING CONDITIONS
Understanding protein dynamics is important for the development of effective drugs targeting muscle wasting conditions. As discussed above, loss of muscle mass can occur as the result of various permutations of protein turnover in which the rate of protein breakdown is greater than protein synthesis, independent of their absolute rates. Depending on the primary side of the balance equation that has been altered to cause loss of muscle mass, a molecular targeting approach for drug development must be considered accordingly. For example, if the primary cause of a catabolic response is the attenuation of protein synthesis, three major targets exist for drug development: 1) implicated signaling pathways [
23], 2) translational processes (i.e., translational capacity and efficiency) [
24], and the availability of precursor AAs [
25]. Although signaling pathways and translational processes are important, protein synthesis is dependent on adequate precursor AAs in the intramuscular fluid compartment, particularly essential AAs (EAAs), which are not endogenously produced. The response to anabolic stimuli, such as resistance exercise and anabolic hormones [
15,
26], may be limited by precursor availability, thus ensuring that an adequate precursor availability is necessary to obtain the desired results. Therefore, we feel that efforts to discover drugs that target muscle wasting will not likely result in clinically meaningful results without understanding the nature of protein turnover. Consistent with this concept, clinical studies have not shown a significant improvement in muscle mass and/or strength by targeting only the molecular pathways of protein synthesis or breakdown (e.g., myostatin inhibitors, leucine supplementation, etc.) without increasing EAA precursor availability [
27-
29]. Conversely, if a catabolic response is primarily caused by accelerated protein breakdown, the loss of muscle mass may theoretically be effectively counteracted by inactivating the responsible molecular pathways because “exogenous precursors” are not required in contrast to protein synthesis (i.e., EAAs). Thus, determining the primary problem regarding protein turnover is critical for development of “effective” drugs to treat muscle wasting diseases. Therefore, the basic principles of stable isotope tracer methodology are reviewed to determine the characteristics of the proteome in vivo, which are applicable to humans, animals, and in vitro models, with emphasis on human skeletal muscle protein dynamics.
STABLE ISOTOPE TRACER METHODS ASSESSING MUSCLE DYNAMICS
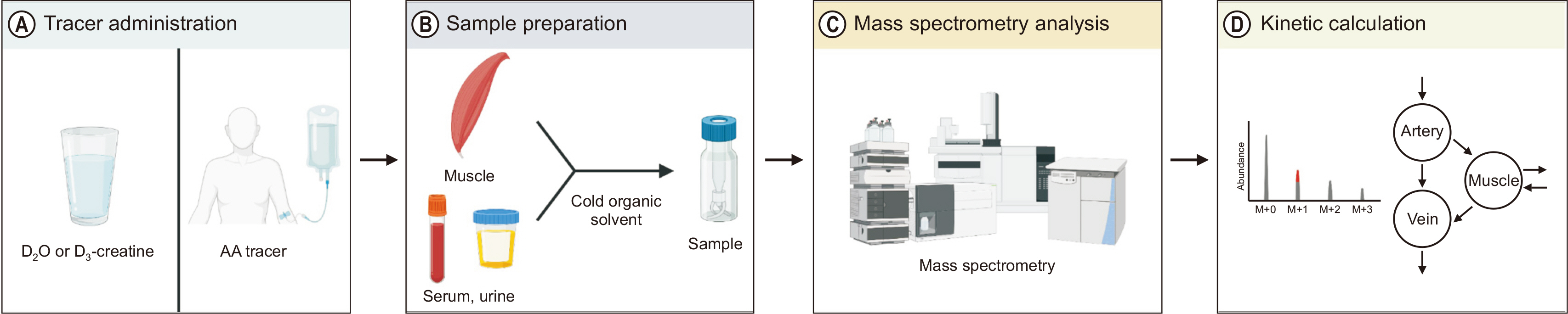
In vivo information on protein dynamics can be assessed using various tracer methods;
Fig. 1 shows analysis using stable isotope-based tracing. Numerous tracer methods exist; however, all are fundamentally based on two tracer models, i.e., tracer dilution and tracer incorporation. The models can be used either independently or in combination to assess in vivo kinetics [
30-
32]. Various aspects of quantitative metabolic fluxes of nearly every biological compound kinetics including glucose, fatty acids, glycerol, and various AAs can be dissected at cellular, organ, and systemic levels using these state-of-the-art methodologies. Several representative methods use either AA tracers (EAAs such as leucine [Leu] or phenylalanine [Phe] tracer are popular because they are not produced endogenously) or D
2O, both of which trace naturally occurring tracee AAs. Both tracer methods employ a stable isotopically labeled compound (either AA tracer or heavy water) in which one or more heavier isotopes (
13C,
2H, or
15N isotopes in AA tracers and
2H isotope in D
2O method) are incorporated somewhere in the tracer compound [
31,
33,
34]. In the following section, the basics of tracer methodology are reviewed and representative tracer methods that enable determination of protein dynamics in vivo evaluated. For more comprehensive information, please refer to the following references [
30,
31,
33,
35,
36] or to the annual workshop on isotope tracer methodology sponsored by the National Institutes of Health – Mouse Metabolic Phenotyping Center (
https://www.mmpc.org/shared/tracers.aspx).
MUSCLE FRACTIONAL SYNTHESIS AND BREAKDOWN
Determination of protein dynamics in vivo includes either 1) administration of a stable isotope AA tracer into the system, typically intravenously (or bolus injection, which is not considered in the present review) or 2) administration (i.p. injection into animal models or oral consumption in humans) of heavy water which labels precursor AAs with deuterium. In both cases, “labeled” AAs are incorporated into product proteins used in conjunction with precursor enrichment determined by mass spectrometry (MS) over the defined experimental period for determination of protein kinetics in muscle and other tissues.
DETERMINATION OF MUSCLE PROTEIN KINETICS USING AN AMINO ACID TRACER INFUSION METHOD
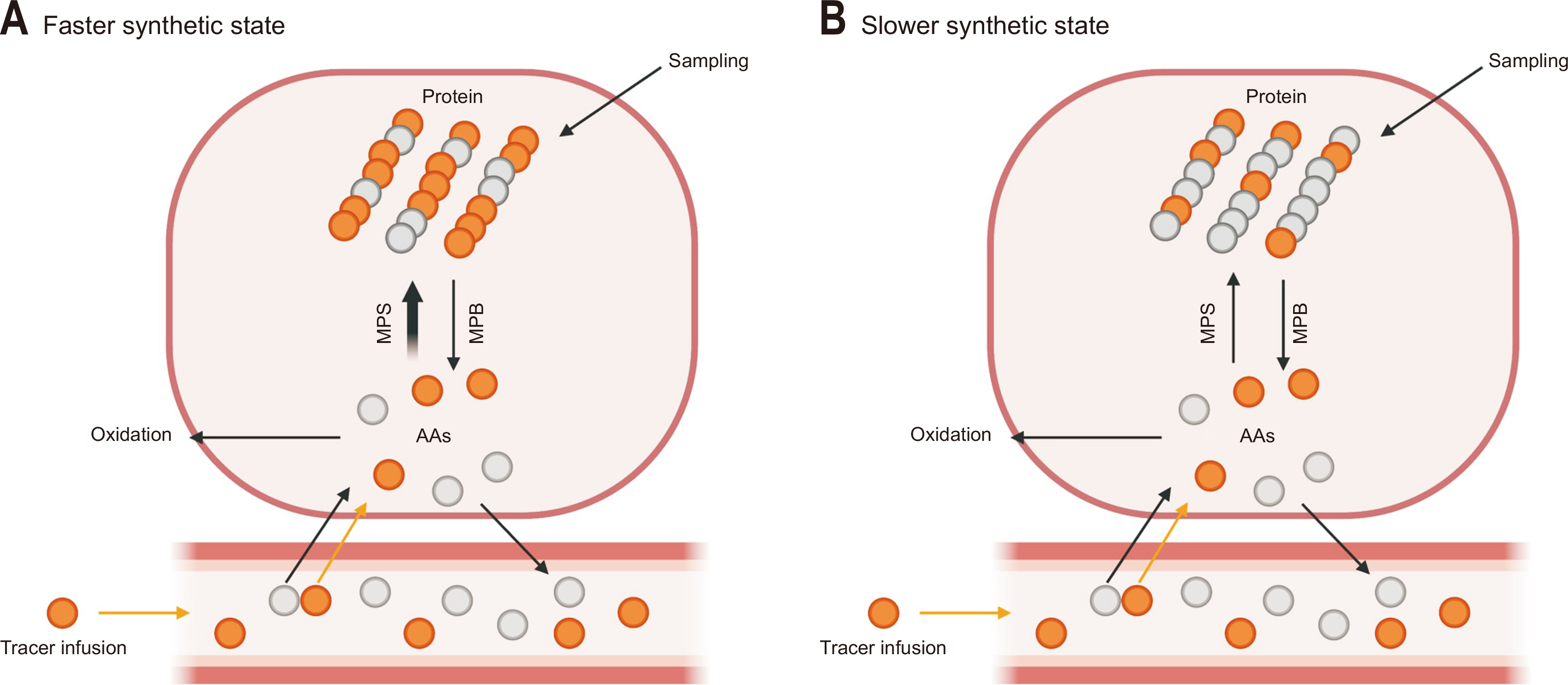
Muscle protein synthesis (MPS) and muscle protein breakdown (MPB) can be assessed using AA tracers and typically determined as the fractional synthetic rate (FSR) and fractional breakdown rate (FBR). The assessment of muscle protein FSR is predicated on the incorporation of precursor (AA) tracer into product (e.g., protein) tracer incorporation approach, which is commonly used to calculate synthesis of any polymers such as DNA [
37,
38], protein [
39,
40], glucose (i.e., gluconeogenesis and oxidation) [
41-
43], fatty acids (i.e., de novo lipogenesis and oxidation) [
14,
44], and triglycerides [
45]. Since AAs are used as building blocks of proteins, the administration of any AA tracer (e.g.,
2H
5-phenylalanine, or 1-
13C-leucine) as a bolus or constant infusion measures the incorporation of the AAs into muscle protein over time (
Fig. 2). When assessing FBR, proteins are considered precursors that release AAs from the process of protein breakdown. Based on this principle, the FBR in muscle can be assessed using the 3-pool A-V tracer model [
46], administration of L-[3-methyl-
2H
3] histidine [
47], or decay of the bound enrichment method [
48]. When comparing FSR and FBR, the pool size (i.e., muscle protein mass) should be considered because FSR and FBR represent the fraction of the pool size that has been newly produced in a defined experimental period. In general, FSR can be directly compared in acute study as the muscle pool size is virtually unchanged, but when the pool size is significantly altered (e.g., muscle hypertrophy, or wasting conditions), the absolute MPS and MPB should be determined to compare effects between groups (absolute MPS or MPB=FSR or FBR×pool size÷100). Detailed information regarding the MPS and MPB using substrate tracers is available elsewhere [
31,
33,
40].
Fig. 2 shows the basic principles of muscle protein FSR and FBR assessment.
HEAVY WATER LABELING METHOD
Heavy water labeling is another tracer method used to determine muscle protein FSR with several advantages. Cumulative muscle protein FSR can be assessed over prolonged periods of time under free-living conditions with simultaneous FSR assessment of different polymers with no apparent true precursor enrichment issue over the traditionally used AA tracer infusion method (e.g.,
13C,
15N, or
2H labeled AA) [
34,
49,
50].
The basic principle of the heavy water labeling method is depicted in
Fig. 3. FSR can be assessed via oral administration in humans [
34] or in small animals such as mice [
51] and rats [
7]. A rapid equilibrium occurs at a certain level of body water enrichment (animals: ~5% MPE [
52], humans: ~0.5% MPE [
34]) which generates monodeuterated water (
2H
1HO) via exchange of hydrogen (
1H or
2H) between water and precursor AAs through trans-amination reactions [
34] and subsequently incorporated into proteins. Finally, muscle protein FSR can be calculated as the change (increase) in enrichment of specific AAs (e.g., alanine with four labeling sites) bound to proteins divided by the precursor (AA) enrichment (directly reflected by water enrichment) because a fixed enrichment ratio exists between water and specific AAs. The enrichment of water or muscle tissue is measured using gas or liquid chromatography-MS [
34]. Regarding FBR measurements, since the changes in muscle mass (i.e., protein pool size) are the cumulated difference between MPS (FSR×pool size) and MPB (FBR×pool size), the integrated MPB over time can be deduced by subtracting the cumulated MPS from the changes in muscle mass.
VIRTUAL BIOPSY: MINIMALLY INVASIVE ASSESSMENT OF MUSCLE PROTEIN KINETICS
The rate of protein turnover measured from muscle samples can serve as a key diagnostic marker for skeletal muscle health [
32]. However, collection of muscle samples via needle biopsy is quite invasive and hinders human participation in research, particularly for children, women, or older adults [
53,
54]. For example, obtaining sufficient muscle tissue in older adults is inherently difficult due to greatly reduced muscle mass (i.e., sarcopenia) [
40]. Therefore, a “virtual biopsy” method provides a solution to this issue [
7,
55] and can be used to quantify muscle protein FSR from blood [
7] or urine [
9] samples (
Fig. 3). The method consists of deuterium labeling of muscle proteins through ingestion of a small amount (100 mL) of heavy water daily for a few days to several months, and measuring muscle-derived proteins from blood (e.g., muscle creatine kinase, carbonic anhydrase 3 isoform) [
7] or urine (e.g., muscle-specific contractile proteins such as titin and myosin light chain 1/3) [
9], which can accurately reflect FSR of muscle proteins in normal heathy individuals (adolescents or adults who do or do not exercise) [
7] and patients such as individuals with Duchenne muscular dystrophy [
9] or chronic liver disease [
56].
WHOLE-BODY AMINO ACID AND PROTEIN KINETICS
Determining protein kinetics at the whole-body level is important for several reasons. First, determination of protein kinetics in muscle may underestimate total protein dynamics, particularly when assessing the overall maximal total anabolic response to dietary protein because a significant portion of protein turnover occurs in other tissues such as rapidly turning-over tissue like the gut, which can later affect muscle protein kinetics [
57]. Second, determination of muscle protein FBR has technical difficulties such as complicated mathematical equations and experimental protocols [
33,
46,
58]. Therefore, determining the whole-body rate of protein breakdown can be a surrogate measure for muscle protein FBR. Furthermore, simultaneous determination of MPS and MPB is impossible within the same time period [
33]. Finally, whole-body protein kinetics can be determined relatively noninvasively from blood samples without sampling muscle tissues [
31,
33]. In the following section, representative tracer methods to determine whole-body protein kinetics using Leu tracer method or dual phenylalanine-tyrosine (Phe-Tyr) are reviewed. Furthermore, we will discuss two approaches to assess postprandial whole-body protein kinetics: digestibility method and intrinsically labeled protein method.
GENERAL PRINCIPLES: ESSENTIAL AMINO ACID TRACER APPROACH
The basic principle for determining whole-body protein kinetics is depicted in
Fig. 4. The general approach to assess in vivo whole-body protein kinetics is based on determining the rate of appearance (
Ra) into and rate of disappearance (
Rd) from plasma of a representative EAA, particularly in the Leu and Phe-Tyr methods. The
Ra (=
Rd in a physiologically steady state) of the unlabeled EAA (tracee) estimated from the EAA tracer based on the dilution model directly reflects the rate of protein breakdown in the fasted state since there is no endogenous production of EAAs (e.g., Leu). The determination of whole-body protein kinetics is predicated on a most complex tracer model which both tracer dilution and incorporation models are integrated with multiple pools and precursors [
30,
59]. For example, in physiological steady states,
Rd of Leu (e.g., of EAA) is equal to
Ra of Leu, the latter of which reflects protein breakdown [
36] and has two metabolic fates: rate of 1) protein synthesis (
Rd2) and 2) oxidation (
Rd3). Protein breakdown cannot be directly determined but deduced from protein synthesis and oxidation (i.e., protein breakdown=protein synthesis–oxidation), the latter of which (i.e., Leu oxidation) can be quantified based on the information of labeled CO
2 enrichment in expired air,
Rd Leu, and infusion rate (F) of
13C labeled Leu. Actual protein synthesis and protein breakdown, thus, net protein balance (protein synthesis–protein breakdown) can be calculated by dividing EAA kinetics by its fractional contribution to protein (e.g., 4% in the case of Phe) [
60,
61]. Compared with the Leu method, the Phe-Tyr method determines hydroxylation of Phe to Tyr, typically from blood, but not oxidation, consequently eliminating the requirement of isotope ratio mass spectrometry for measuring expired CO
2 enrichment [
62]. For the postprandial assessment of whole-body protein kinetics, the Phe-Tyr method is further discussed due to our extensive experience with this method [
63,
64].
POSTPRANDIAL ASSESSMENT
Whole-body protein kinetics can be determined in the non-steady state (e.g., following consumption of a meal containing proteins or AAs). The appearance of ingested tracee into the peripheral blood must also be quantified to calculate the rate of protein breakdown in the fed state. The postprandial assessment can be accomplished by adding the chemically identical but differently labeled tracer AA from the tracer infused intravenously to the protein meal. Two methods have been widely used: 1) intrinsically labeled protein method [
65] and 2) bioavailability method [
66,
67]. In the intrinsically labeled protein method, research participants consume labeled protein (labeled AAs in the proteins) [
68,
69]. Although in this method, dilution of tracer coming from the labeled proteins assumedly does not occur between the point of the ingestion and the point of blood sampling, this is not the case. This dilution underestimates contribution of exogenous AAs to peripheral blood (
Ra EXO) and consequently overestimates protein breakdown [
36,
61] resulting in underestimation of net protein balance: Total
Ra (
Ra TOT)=
Ra endogenous (
Ra ENDO, reflecting protein breakdown)+
Ra EXO (AAs from food). When solving for
Ra ENDO, protein breakdown=
Ra TOT–
Ra EXO. In the bioavailability method,
Ra EXO is estimated by correcting the total protein consumption for true ileal digestibility [
61,
70]. The method assumes no gain or loss of intestinal protein in the process of digestion and absorption, which may in turn introduce errors, which would be random but not accompanied with any systematic bias in the calculated protein kinetics [
61,
70].
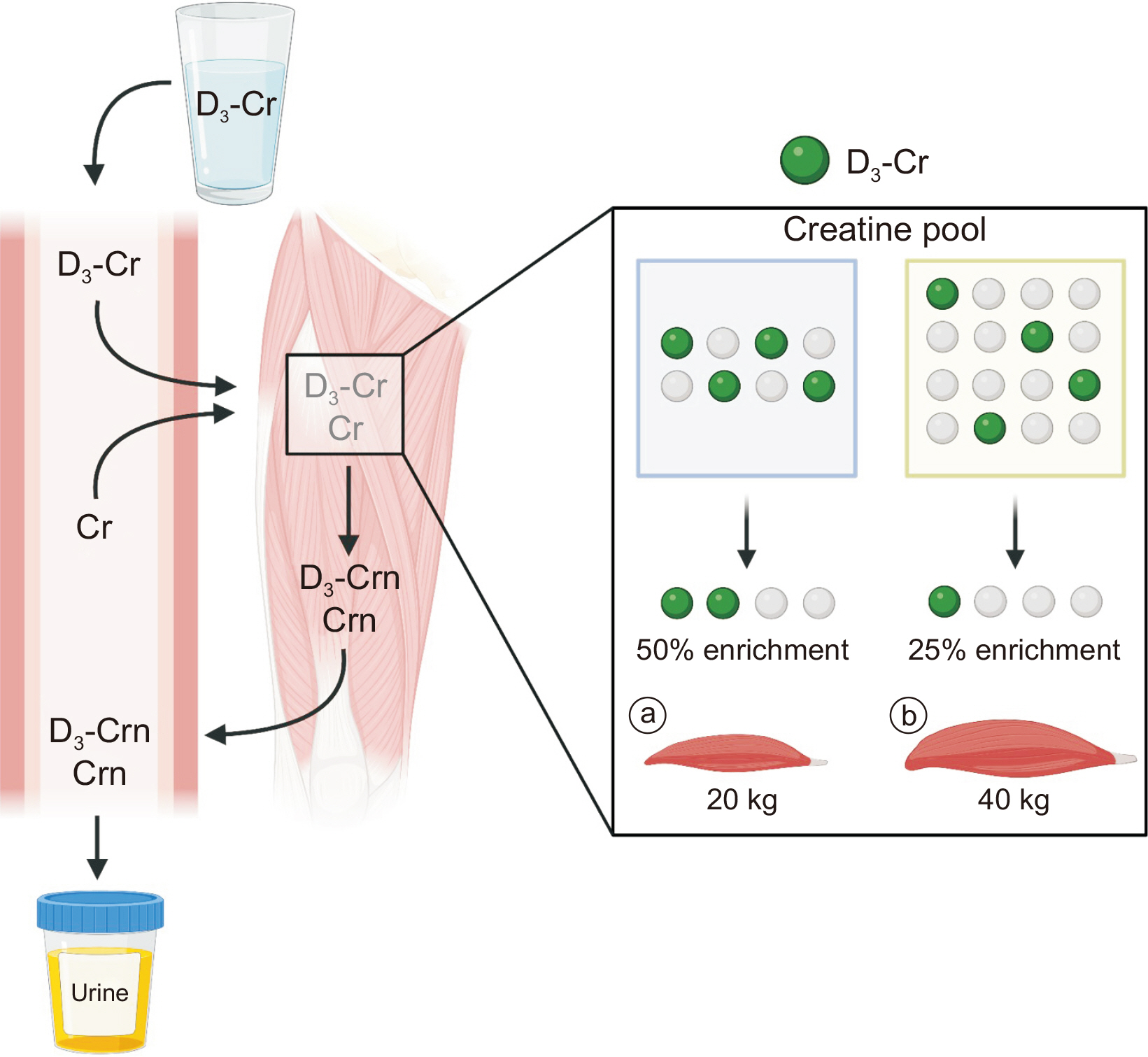
ASSESSMENT OF DIRECT MUSCLE MASS: D3-CREATINE DILUTION METHOD
Knowledge regarding the dynamic nature of muscle proteins (“kinetics”) is important as well as determining direct muscle mass (changes) to examine the efficacy of new therapeutics (nutrition, exercise, or drug) for muscle, and can be accomplished with the recently developed non-invasive method, called the D
3-creatine dilution method (
Fig. 5). The method is predicated based on several assumptions: orally consumed D
3-creatine is 1) completely absorbed; 2) remains in the body, more specifically in skeletal muscle; and 3) skeletal muscle creatine concentration is consistent [
71-
73]. These assumptions are arguably valid [
40] although some controversy has recently been suggested [
74], which should be examined more carefully in the future. Briefly, an orally consumed small dose of D
3-creatine will be diluted as the direct function of the abundance of existing creatine pool, which reflects muscle mass since creatine concentration is constant and resides mostly in skeletal muscle [
55,
71]. In addition, a constant irreversible conversion from creatine (both labeled and unlabeled) to creatinine (preservation of the labeled to unlabeled ratio) exists and excreted into urine in accordance with their respective abundance [
55]. Based on the ratio between creatine and creatinine, the total creatine pool size can be obtained by dividing the dose of D
3-creatine by the urine D
3-creatinine enrichment. The skeletal muscle mass can be estimated by dividing the known creatine pool size (4.3 g/kg) by the creatine concentration in the muscle [
9].
CONCLUSION
Skeletal muscle protein is in a constant state of turnover (i.e., rates of protein synthesis and breakdown). Although protein turnover is in a balanced state (synthesis=breakdown) in normal healthy individuals over time, a prolonged imbalance of muscle protein turnover (breakdown>synthesis) can lead to muscle wasting conditions such as sarcopenia and cachexia. Loss of muscle mass has been a target of drug development; however, this effort has not had clinical success to date, possibly due to a lack of understanding the dynamic nature of the muscle proteome. Stable isotope tracer methodology enables quantification of protein dynamics (i.e., kinetics) in humans and animals as well as in in vitro models. In conjunction with molecular and cellular biology tools, stable isotope tracer methodology provides an in-depth assessment of dynamic changes in the proteome as well as simultaneous understanding of the molecular mechanisms for the observed kinetics of proteome, allowing researchers to develop effective therapeutic strategies (e.g., nutrition, exercise, and drugs) to treat muscle wasting diseases.
AUTHOR CONTRIBUTIONS
Conceptualization: IYK. Funding acquisition: IYK, SP, JJ. Visualization: HJK. Writing – original draft: IYK. Writing – review & editing: IYK, SP, JJ, YK, HJK.
CONFLICTS OF INTEREST
IYK and SP are shareholders in Myocare, Inc. and Myocro, Inc. The other authors declare no competing interests.
FUNDING
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2021R1A2C3005801), the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A3A13071529), the Brain Pool Program funded by the Ministry of Science and ICT through the National Research Foundation of Korea (2019H1D3A1A01071043), and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1I1A1A01074380).
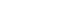
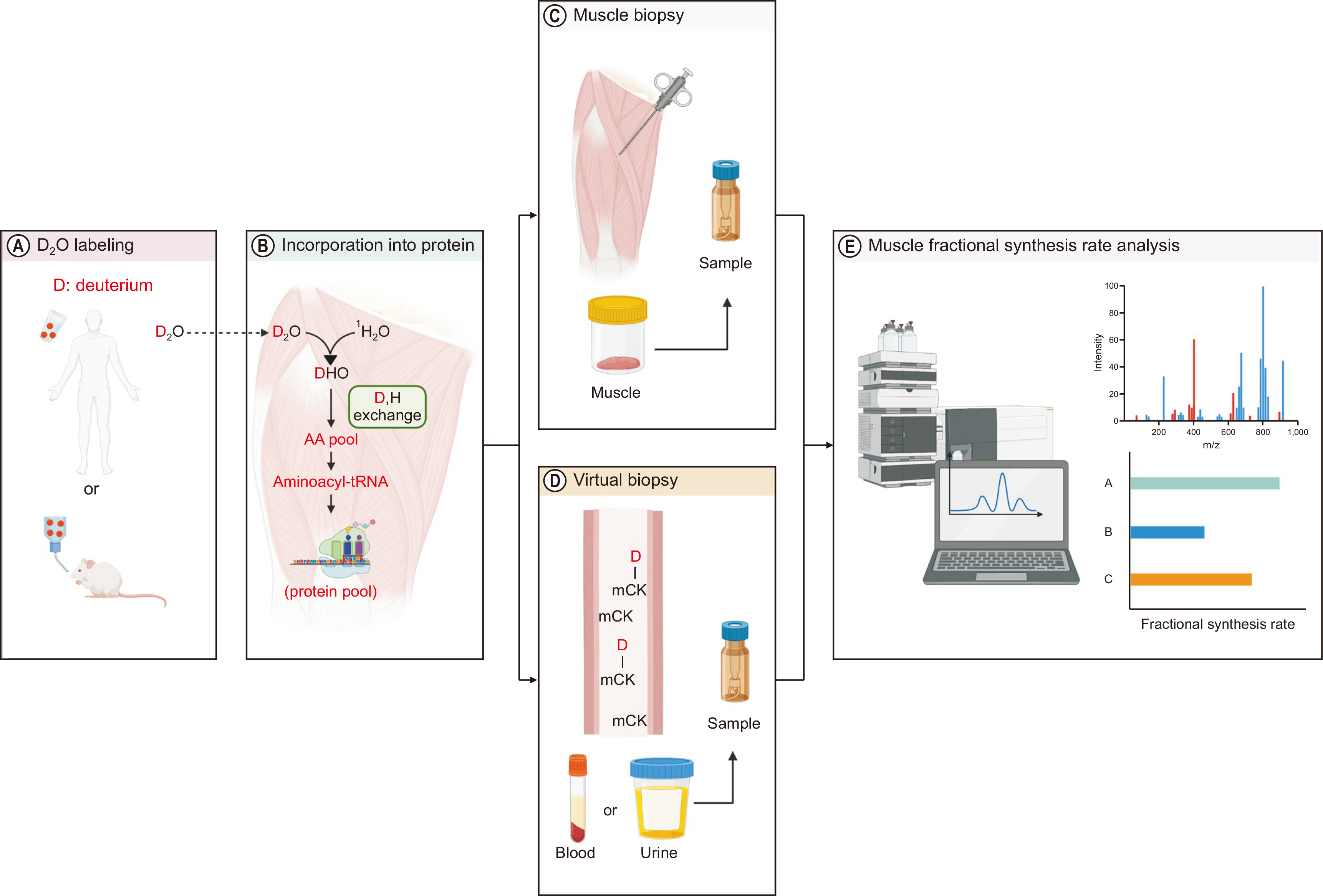
Fig. 1
Schematic diagram of stable isotope tracer-based muscle metabolic research. (A) One or more of tracers are administered into the circulation typically via intravenous infusion or oral bolus to assess muscle protein dynamics. (B, C) Samples are processed for mass spectrometry analysis of tracer enrichment of water, free amino acids (AAs) or AAs bound to proteins. (D) Finally, tracee kinetics are mathematically calculated based on the dose information of the rate of tracer administered and measured enrichment. This figure was created with BioRender.com.
D2O = Deuterium oxide.
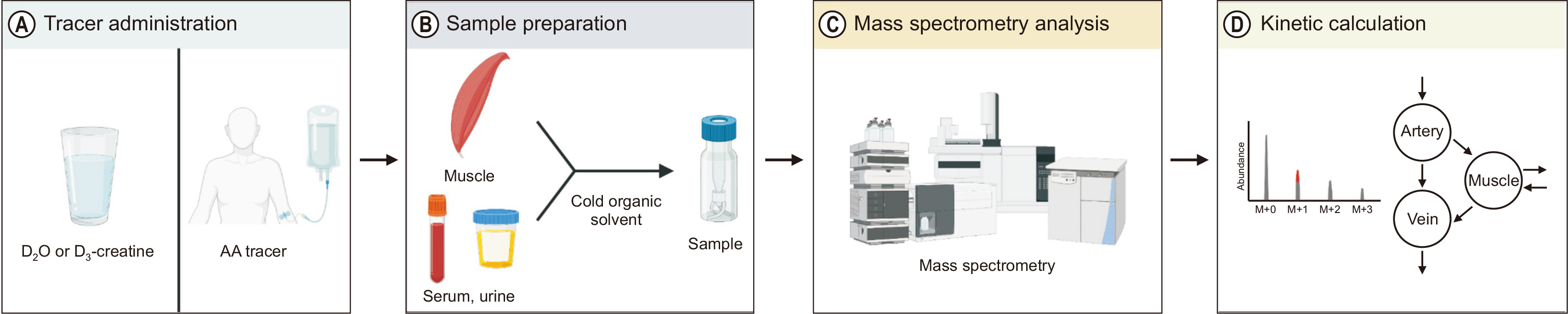
Fig. 2
Assessments of muscle protein fractional synthetic rate using amino acid (AA) tracer method. Both labeled tracer and unlabeled AAs cross the muscle cell membrane in proportion to their relative concentrations (i.e., tracer-to-tracee ratio) to enter the free AA precursor pool from where they are either oxidized or incorporated into proteins. For a given precursor enrichment, a greater labeling of product (i.e., protein enrichment) is achieved in the faster synthesis condition (A) compared with the slower synthesis condition (B). This figure was created with BioRender.com.
MPS = muscle protein synthesis; MPB = muscle protein breakdown.
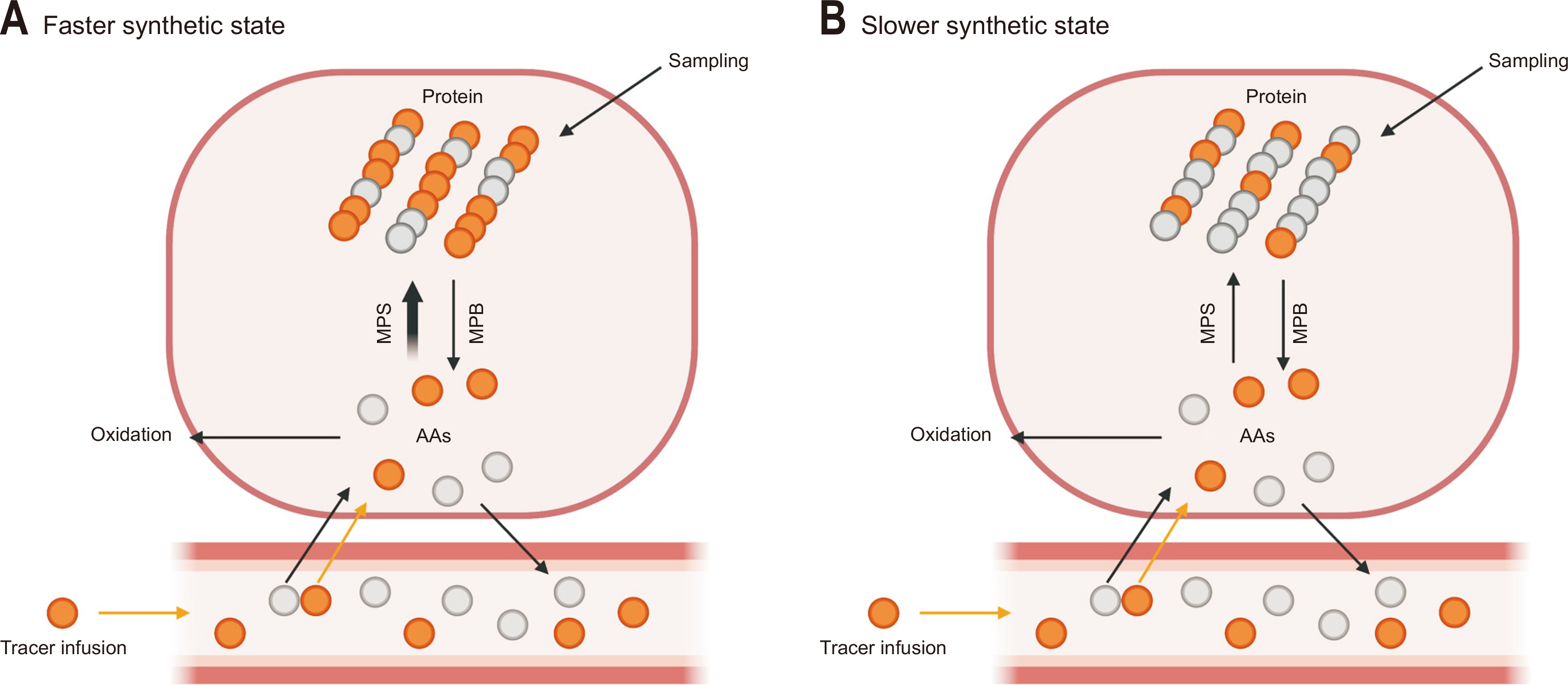
Fig. 3
Assessments of muscle protein fractional synthetic rate (FSR) using heavy water labeling method. (A) A small dose of heavy water (deuterium oxide, D2O) is orally administered to achieve target enrichment (~1% for humans and ~5% for animals) and assess FSR of muscle protein. (B) D2O administered into the skeletal muscle will form mono-deuterated water (i.e., 2H1HO). The 2H will be incorporated into precursors, such as amino acids (AAs). The 2H-labeled AAs are incorporated into new muscle proteins. (C, D) Muscle protein FSR (individual, subgroup, or mixed proteins) is determined based on the information on precursor AA enrichment and changes in product enrichment over a defined time. (E) In addition, enrichment is accessed using gas or liquid chromatography-mass spectrometry in samples obtained via needle biopsy, blood, or urine. This figure was created with BioRender.com.
mCK = muscle creatine kinase.
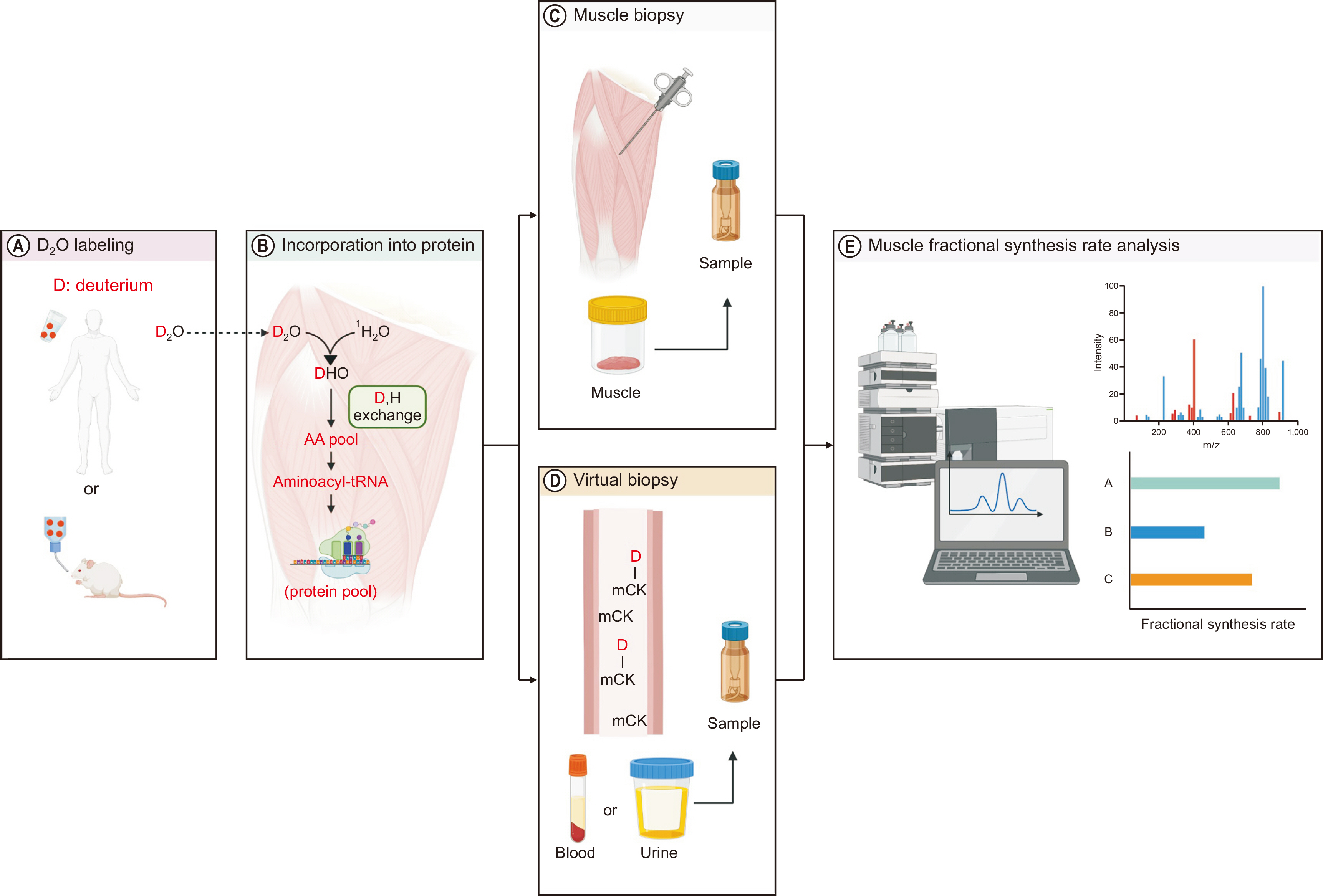
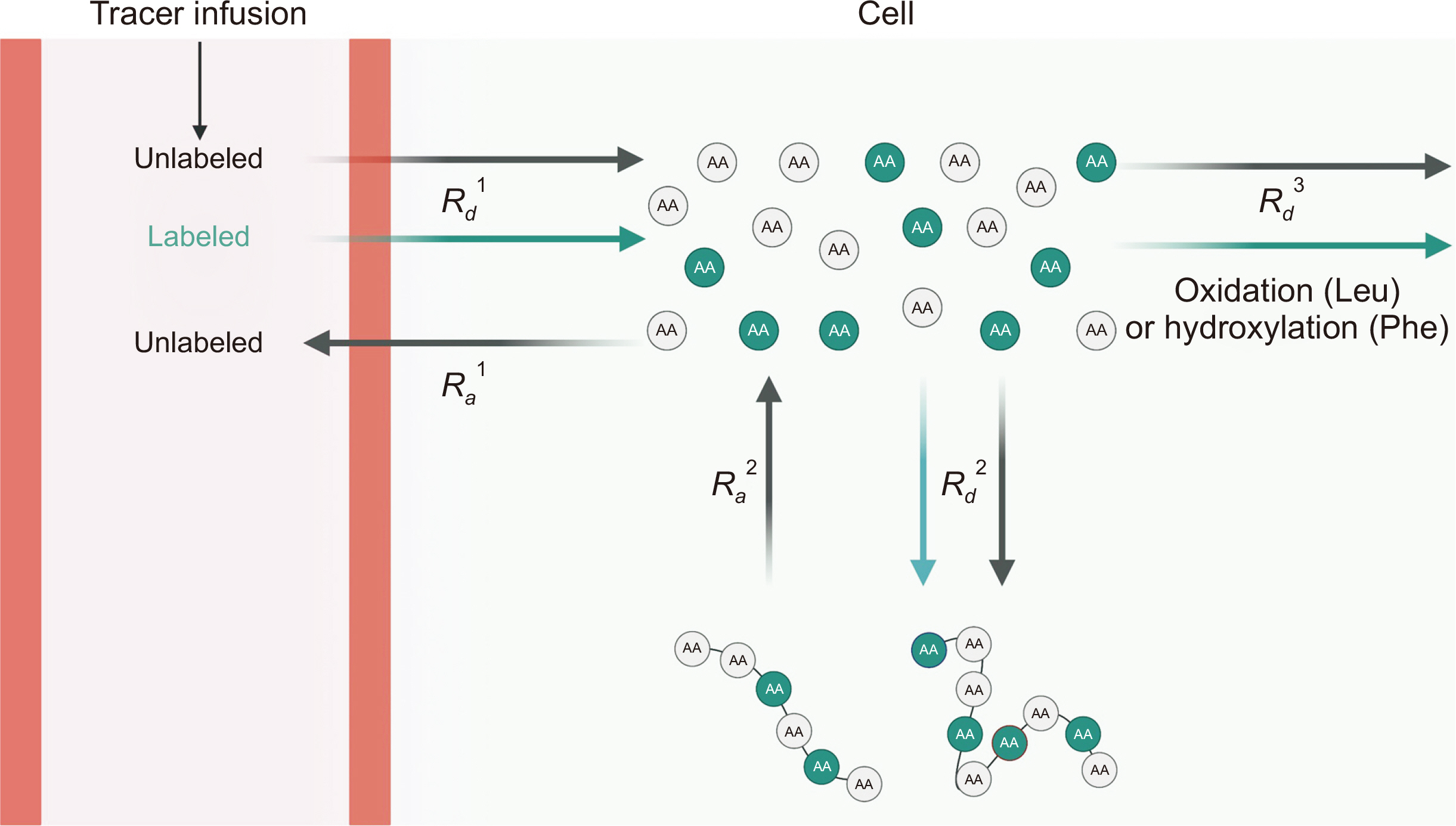
Fig. 4
Assessment of whole-body protein kinetics. Both labeled (from outside of the body via plasma) and unlabeled amino acids (AAs) cross the cell membrane in proportion to their relative concentrations (i.e., tracer-to-tracee ratio) to enter the free intracellular AA precursor pool (i.e., Rd1). Once inside cells, the tracer can be oxidized (or hydroxylated in the case of Phe, Rd3) or incorporated into protein (Rd2). In a physiological steady state, Ra1 reflects the rate of protein breakdown and is equal to Rd1. This figure was created with BioRender.com.
Rd = rate of disappearance; Ra = rate of appearance; Leu = leucine; Phe = phenylalanine.
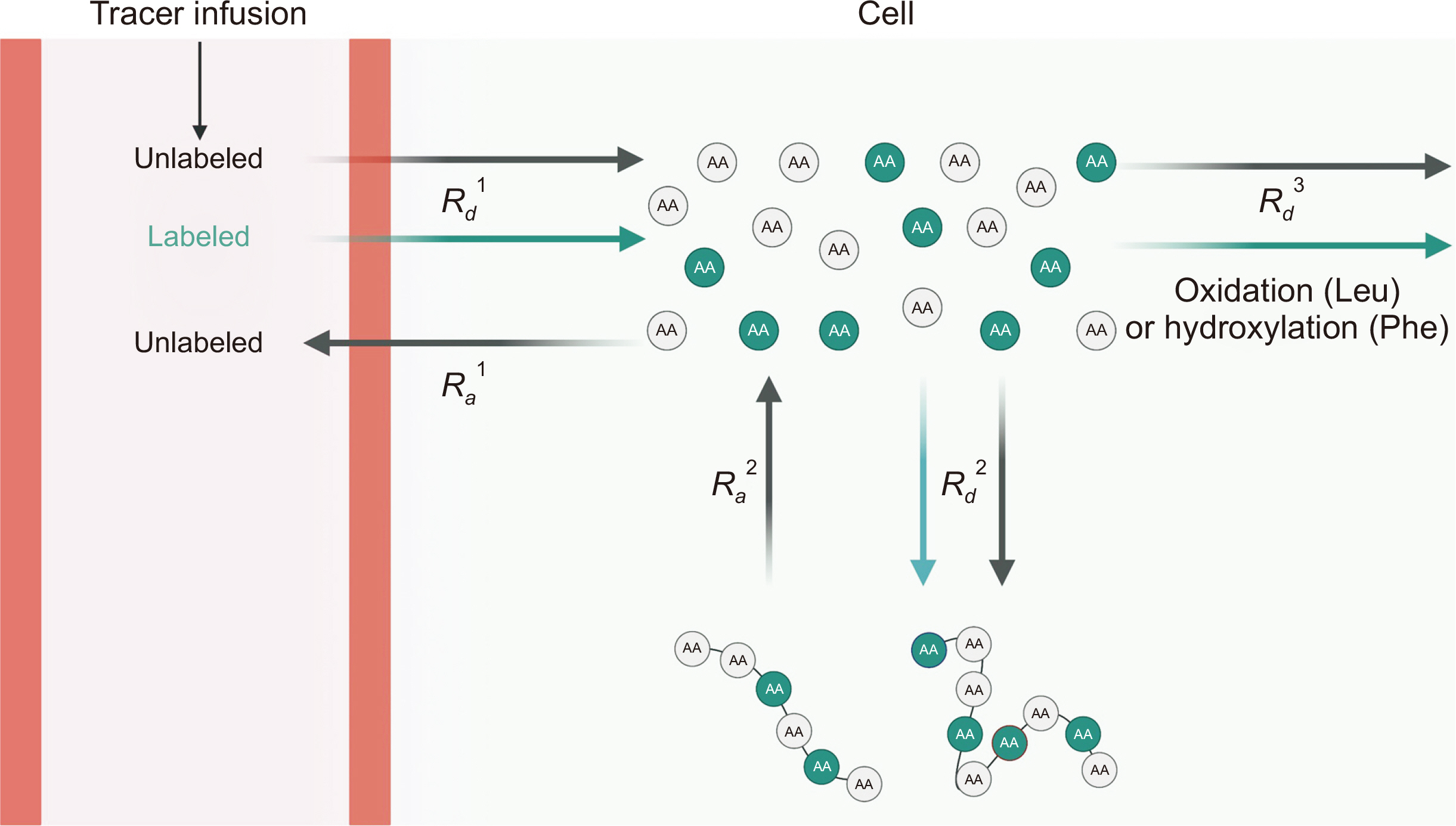
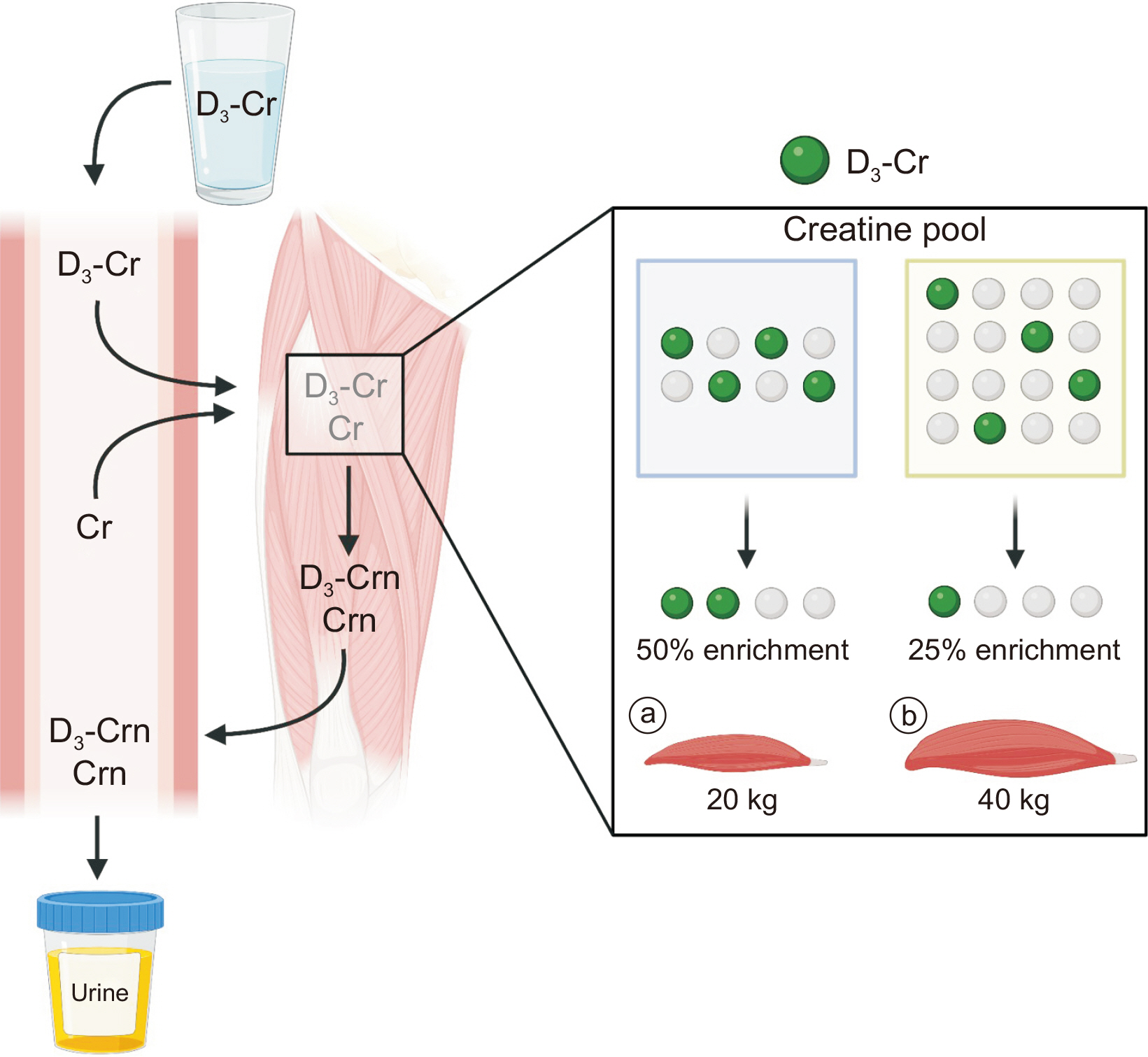
Fig. 5
Direct assessments of whole-body functional muscle mass using D3-Cr dilution method. A small dose of absorbed D3-Cr and endogenous Cr in the skeletal muscle will irreversibly be converted to Crn. The Crn enrichment from urine reflects the dilution of D3-Cr as a function of the abundance of existing creatine pool. Knowledge of 1) whole-body Cr pool size (determined) and 2) muscle Cr concentrations (known to be constant) enables calculations of skeletal muscle mass. For a given D3-Cr oral consumption, a greater dilution of D3-Cr will occur in cases of large muscle mass (a) compared with smaller muscle mass (b). This figure was created with BioRender.com.
Cr = creatine; Crn = creatinine.
References
- 1. Schoenheimer R, Ratner S, Rittenberg D. Studies in protein metabolism: VII. The metabolism of tyrosine. J Biol Chem 1939;127:333-44.
- 2. Schoenheimer R. The dynamic state of body constituents. 2nd ed. Cambridge (MA): Harvard University Express; 1946.
- 3. Biolo G, Fleming RY, Maggi SP, Nguyen TT, Herndon DN, Wolfe RR. Inverse regulation of protein turnover and amino acid transport in skeletal muscle of hypercatabolic patients. J Clin Endocrinol Metab 2002;87:3378-84. ArticlePubMed
- 4. Bukhari SS, Phillips BE, Wilkinson DJ, Limb MC, Rankin D, Mitchell WK, et al. Intake of low-dose leucine-rich essential amino acids stimulates muscle anabolism equivalently to bolus whey protein in older women at rest and after exercise. Am J Physiol Endocrinol Metab 2015;308:E1056-65. ArticlePubMed
- 5. Neinast MD, Jang C, Hui S, Murashige DS, Chu Q, Morscher RJ, et al. Quantitative analysis of the whole-body metabolic fate of branched-chain amino acids. Cell Metab 2019;29:417-29.e4. ArticlePubMedPMC
- 6. Hines KM, Ford GC, Klaus KA, Irving BA, Ford BL, Johnson KL, et al. Application of high-resolution mass spectrometry to measure low abundance isotope enrichment in individual muscle proteins. Anal Bioanal Chem 2015;407:4045-52. ArticlePubMedPMCPDF
- 7. Shankaran M, King CL, Angel TE, Holmes WE, Li KW, Colangelo M, et al. Circulating protein synthesis rates reveal skeletal muscle proteome dynamics. J Clin Invest 2016;126:288-302. ArticlePubMedPMC
- 8. Murphy CH, Shankaran M, Churchward-Venne TA, Mitchell CJ, Kolar NM, Burke LM, et al. Effect of resistance training and protein intake pattern on myofibrillar protein synthesis and proteome kinetics in older men in energy restriction. J Physiol 2018;596:2091-120. ArticlePubMedPMCPDF
- 9. Evans WJ, Shankaran M, Smith EC, Morris C, Nyangau E, Bizieff A, et al. Profoundly lower muscle mass and rate of contractile protein synthesis in boys with Duchenne muscular dystrophy. J Physiol 2021;599:5215-27. ArticlePubMedPDF
- 10. Burd NA, West DW, Staples AW, Atherton PJ, Baker JM, Moore DR, et al. Low-load high volume resistance exercise stimulates muscle protein synthesis more than high-load low volume resistance exercise in young men. PLoS One 2010;5:e12033. ArticlePubMedPMC
- 11. Wilkinson SB, Phillips SM, Atherton PJ, Patel R, Yarasheski KE, Tarnopolsky MA, et al. 2008;Differential effects of resistance and endurance exercise in the fed state on signalling molecule phosphorylation and protein synthesis in human muscle. J Physiol 586:3701-17. ArticlePubMedPMC
- 12. Katsanos CS, Kobayashi H, Sheffield-Moore M, Aarsland A, Wolfe RR. A high proportion of leucine is required for optimal stimulation of the rate of muscle protein synthesis by essential amino acids in the elderly. Am J Physiol Endocrinol Metab 2006;291:E381-7. ArticlePubMed
- 13. Strawford A, Antelo F, Christiansen M, Hellerstein MK. Adipose tissue triglyceride turnover, de novo lipogenesis, and cell proliferation in humans measured with 2H2O. Am J Physiol Endocrinol Metab 2004;286:E577-88. ArticlePubMed
- 14. Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, Parks EJ. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest 2005;115:1343-51. ArticlePubMedPMC
- 15. Kim IY, Schutzler S, Schrader A, Spencer H, Kortebein P, Deutz NE, et al. Quantity of dietary protein intake, but not pattern of intake, affects net protein balance primarily through differences in protein synthesis in older adults. Am J Physiol Endocrinol Metab 2015;308:E21-8. ArticlePubMedPMC
- 16. Kim IY, Schutzler S, Schrader A, Spencer HJ, Azhar G, Ferrando AA, et al. The anabolic response to a meal containing different amounts of protein is not limited by the maximal stimulation of protein synthesis in healthy young adults. Am J Physiol Endocrinol Metab 2016;310:E73-80. ArticlePubMedPMC
- 17. Kim IY, Shin YA, Schutzler SE, Azhar G, Wolfe RR, Ferrando AA. Quality of meal protein determines anabolic response in older adults. Clin Nutr 2018;37(6 Pt A):2076-2083. ArticlePubMedPMC
- 18. Kim IY, Schutzler S, Schrader AM, Spencer HJ, Azhar G, Wolfe RR, et al. 2018;Protein intake distribution pattern does not affect anabolic response, lean body mass, muscle strength or function over 8 weeks in older adults: a randomized-controlled trial. Clin Nutr 37:488-93. ArticlePubMedPMC
- 19. Park SW, Goodpaster BH, Strotmeyer ES, de Rekeneire N, Harris TB, Schwartz AV, et al. Decreased muscle strength and quality in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes 2006;55:1813-8. PubMed
- 20. Fitts RH, Romatowski JG, Peters JR, Paddon-Jones D, Wolfe RR, Ferrando AA. The deleterious effects of bed rest on human skeletal muscle fibers are exacerbated by hypercortisolemia and ameliorated by dietary supplementation. Am J Physiol Cell Physiol 2007;293:C313-20. ArticlePubMed
- 21. Robinson MM, Dasari S, Konopka AR, Johnson ML, Manjunatha S, Esponda RR, et al. Enhanced protein translation underlies improved metabolic and physical adaptations to different exercise training modes in young and old humans. Cell Metab 2017;25:581-92. ArticlePubMedPMC
- 22. Perry RJ, Wang Y, Cline GW, Rabin-Court A, Song JD, Dufour S, et al. Leptin mediates a glucose-fatty acid cycle to maintain glucose homeostasis in starvation. Cell 2018;172:234-48.e17. ArticlePubMedPMC
- 23. Kimball SR, Jefferson LS. Control of translation initiation through integration of signals generated by hormones, nutrients, and exercise. J Biol Chem 2010;285:29027-32. ArticlePubMedPMC
- 24. Bamman MM, Roberts BM, Adams GR. Molecular regulation of exercise-induced muscle fiber hypertrophy. Cold Spring Harb Perspect Med 2018;8:a029751. ArticlePubMedPMC
- 25. Kim IY, Deutz NEP, Wolfe RR. 2018;Update on maximal anabolic response to dietary protein. Clin Nutr 37:411-8. ArticlePubMedPMC
- 26. Breen L, Phillips SM. Interactions between exercise and nutrition to prevent muscle waste during ageing. Br J Clin Pharmacol 2013;75:708-15. ArticlePubMedPMCPDF
- 27. Verhoeven S, Vanschoonbeek K, Verdijk LB, Koopman R, Wodzig WK, Dendale P, et al. Long-term leucine supplementation does not increase muscle mass or strength in healthy elderly men. Am J Clin Nutr 2009;89:1468-75. ArticlePubMed
- 28. Leenders M, van Loon LJ. Leucine as a pharmaconutrient to prevent and treat sarcopenia and type 2 diabetes. Nutr Rev 2011;69:675-89. ArticlePubMed
- 29. Saitoh M, Ishida J, Ebner N, Anker SD, Springer J, von Haehling S. Myostatin inhibitors as pharmacological treatment for muscle wasting and muscular dystrophy. JCSM Clin Reports 2017;2:1-10. ArticlePDF
- 30. Kim IY, Park S, Kim Y, Kim HJ, Wolfe RR. Tracing metabolic flux in vivo: basic model structures of tracer methodology. Exp Mol Med 2022;54:1311-22. ArticlePubMedPMCPDF
- 31. Kim IY, Suh SH, Lee IK, Wolfe RR. Applications of stable, nonradioactive isotope tracers in in vivo human metabolic research. Exp Mol Med 2016;48:e203. ArticlePubMedPMCPDF
- 32. Park S, Church DD, Azhar G, Schutzler SE, Ferrando AA, Wolfe RR. 2020;Anabolic response to essential amino acid plus whey protein composition is greater than whey protein alone in young healthy adults. J Int Soc Sports Nutr 17:9.ArticlePubMedPMCPDF
- 33. Wolfe RR, Chinkes DL. Isotope tracers in metabolic research: principles and practice of kinetic analysis. 2nd ed. New York (NY): Wiley; 2004.
- 34. Gasier HG, Fluckey JD, Previs SF. The application of 2H2O to measure skeletal muscle protein synthesis. Nutr Metab (Lond) 2010;7:31.ArticlePubMedPMC
- 35. Cobelli C, Foster D, Toffolo G. Tracer kinetics in biomedical research: from data to model. New York (NY): Springer; 2002.
- 36. Wolfe RR, Kim IY, Park S, Ferrando A. Tracing metabolic flux to assess optimal dietary protein and amino acid consumption. Exp Mol Med 2022;54:1323-31. ArticlePubMedPMCPDF
- 37. Zhang XJ, Chinkes DL, Wu Z, Martini WZ, Wolfe RR. Fractional synthesis rates of DNA and protein in rabbit skin are not correlated. J Nutr 2004;134:2401-6. ArticlePubMed
- 38. Martini WZ, Chinkes DL, Wolfe RR. Quantification of DNA synthesis from different pathways in cultured human fibroblasts and myocytes. Metabolism 2004;53:128-33. ArticlePubMed
- 39. Park S, Jang J, Choi MD, Shin YA, Schutzler S, Azhar G, et al. The anabolic response to dietary protein is not limited by the maximal stimulation of protein synthesis in healthy older adults: a randomized crossover trial. Nutrients 2020;12:3276.ArticlePubMedPMC
- 40. Kim IY, Park S, Jang J, Wolfe RR. Understanding muscle protein dynamics: technical considerations for advancing sarcopenia research. Ann Geriatr Med Res 2020;24:157-65. ArticlePubMedPMCPDF
- 41. Kim IY, Park S, Kim Y, Chang Y, Choi CS, Suh SH, et al. In vivo and in vitro quantification of glucose kinetics: from bedside to bench. Endocrinol Metab (Seoul) 2020;35:733-49. ArticlePubMedPMCPDF
- 42. Siler SQ, Neese RA, Christiansen MP, Hellerstein MK. The inhibition of gluconeogenesis following alcohol in humans. Am J Physiol 1998;275:E897-907. ArticlePubMed
- 43. Coggan AR, Kohrt WM, Spina RJ, Bier DM, Holloszy JO. Endurance training decreases plasma glucose turnover and oxidation during moderate-intensity exercise in men. J Appl Physiol (1985) 1990;68:990-6. ArticlePubMed
- 44. Sidossis LS, Coggan AR, Gastaldelli A, Wolfe RR. Pathway of free fatty acid oxidation in human subjects. Implications for tracer studies. J Clin Invest 1995;95:278-84. ArticlePubMedPMC
- 45. Barrows BR, Timlin MT, Parks EJ. Spillover of dietary fatty acids and use of serum nonesterified fatty acids for the synthesis of VLDL-triacylglycerol under two different feeding regimens. Diabetes 2005;54:2668-73. ArticlePubMedPDF
- 46. Zhang XJ, Chinkes DL, Sakurai Y, Wolfe RR. An isotopic method for measurement of muscle protein fractional breakdown rate in vivo. Am J Physiol 1996;270(5 Pt 1):E759-67. ArticlePubMed
- 47. Vissers YL, von Meyenfeldt MF, Braulio VB, Luiking YC, Deutz NE. 2003;Measuring whole-body actin/myosin protein breakdown in mice using a primed constant stable isotope-infusion protocol. Clin Sci (Lond) 104:585-90. ArticlePubMedPDF
- 48. Tuvdendorj D, Chinkes DL, Herndon DN, Zhang XJ, Wolfe RR. A novel stable isotope tracer method to measure muscle protein fractional breakdown rate during a physiological non-steady-state condition. Am J Physiol Endocrinol Metab 2013;304:E623-30. ArticlePubMedPMC
- 49. Herath K, Bhat G, Miller PL, Wang SP, Kulick A, Andrews-Kelly G, et al. Equilibration of (2)H labeling between body water and free amino acids: enabling studies of proteome synthesis. Anal Biochem 2011;415:197-9. ArticlePubMed
- 50. Price JC, Holmes WE, Li KW, Floreani NA, Neese RA, Turner SM, et al. Measurement of human plasma proteome dynamics with (2)H(2)O and liquid chromatography tandem mass spectrometry. Anal Biochem 2012;420:73-83. ArticlePubMed
- 51. Kim Y, Park S, Lee J, Jang J, Jung J, Koh JH, et al. Essential amino acid-enriched diet alleviates dexamethasone-induced loss of muscle mass and function through stimulation of myofibrillar protein synthesis and improves glucose metabolism in mice. Metabolites 2022;12:84.ArticlePubMedPMC
- 52. Ussing HH. The rate of protein renewal in mice and rats studied by means of heavy hydrogen. Acta Physiol Scand 1941;2:209-21. Article
- 53. Wilson D, Breen L, Lord JM, Sapey E. The challenges of muscle biopsy in a community based geriatric population. BMC Res Notes 2018;11:830.ArticlePubMedPMCPDF
- 54. Verhaart IEC, Johnson A, Thakrar S, Vroom E, De Angelis F, Muntoni F, et al. Muscle biopsies in clinical trials for Duchenne muscular dystrophy - patients' and caregivers' perspective. Neuromuscul Disord 2019;29:576-84. ArticlePubMed
- 55. Hellerstein M, Evans W. Recent advances for measurement of protein synthesis rates, use of the 'virtual biopsy' approach, and measurement of muscle mass. Curr Opin Clin Nutr Metab Care 2017;20:191-200. ArticlePubMed
- 56. Decaris ML, Emson CL, Li K, Gatmaitan M, Luo F, Cattin J, et al. Turnover rates of hepatic collagen and circulating collagen-associated proteins in humans with chronic liver disease. PLoS One 2015;10:e0123311. ArticlePubMedPMC
- 57. Deutz NE, Wolfe RR. 2013;Is there a maximal anabolic response to protein intake with a meal? Clin Nutr 32:309-13. ArticlePubMedPMC
- 58. Zhang XJ, Chinkes DL, Wolfe RR. Measurement of muscle protein fractional synthesis and breakdown rates from a pulse tracer injection. Am J Physiol Endocrinol Metab 2002;283:E753-64. ArticlePubMed
- 59. Previs SF, Brunengraber H. Methods for measuring gluconeogenesis in vivo. Curr Opin Clin Nutr Metab Care 1998;1:461-5. ArticlePubMed
- 60. Nair KS, Halliday D, Griggs RC. Leucine incorporation into mixed skeletal muscle protein in humans. Am J Physiol 1988;254(2 Pt 1):E208-13. ArticlePubMed
- 61. Wolfe RR, Park S, Kim IY, Moughan PJ, Ferrando AA. 2020;Advances in stable isotope tracer methodology part 2: new thoughts about an "old" method-measurement of whole body protein synthesis and breakdown in the fed state. J Investig Med 68:11-5. ArticlePubMedPDF
- 62. Carraro F, Stuart CA, Hartl WH, Rosenblatt J, Wolfe RR. Effect of exercise and recovery on muscle protein synthesis in human subjects. Am J Physiol 1990;259(4 Pt 1):E470-6. ArticlePubMed
- 63. Kim IY, Park S, Smeets ETHC, Schutzler S, Azhar G, Wei JY, et al. 2019;Consumption of a specially-formulated mixture of essential amino acids promotes gain in whole-body protein to a greater extent than a complete meal replacement in older women with heart failure. Nutrients 11:1360.ArticlePubMedPMC
- 64. Park S, Church DD, Schutzler SE, Azhar G, Kim IY, Ferrando AA, et al. Metabolic evaluation of the dietary guidelines' ounce equivalents of protein food sources in young adults: a randomized controlled trial. J Nutr 2021;151:1190-6. ArticlePubMedPMCPDF
- 65. van Loon LJ, Boirie Y, Gijsen AP, Fauquant J, de Roos AL, Kies AK, et al. The production of intrinsically labeled milk protein provides a functional tool for human nutrition research. J Dairy Sci 2009;92:4812-22. ArticlePubMed
- 66. Wolfe RR, Kim IY, Church DD, Moughan PJ, Park S, Ferrando AA. Whole-body protein kinetic models to quantify the anabolic response to dietary protein consumption. Clin Nutr Open Sci 2021;36:78-90. Article
- 67. Fuller MF, Tomé D. In vivo determination of amino acid bioavailability in humans and model animals. J AOAC Int 2005;88:923-34. ArticlePubMedPDF
- 68. Metges CC, El-Khoury AE, Selvaraj AB, Tsay RH, Atkinson A, Regan MM, et al. Kinetics of L-[1-(13)C]leucine when ingested with free amino acids, unlabeled or intrinsically labeled casein. Am J Physiol Endocrinol Metab 2000;278:E1000-9. PubMed
- 69. Trommelen J, Kouw IWK, Holwerda AM, Snijders T, Halson SL, Rollo I, et al. Presleep dietary protein-derived amino acids are incorporated in myofibrillar protein during postexercise overnight recovery. Am J Physiol Endocrinol Metab 2018;314:E457-67. ArticlePubMed
- 70. Rutherfurd SM, Moughan PJ. The digestible amino acid composition of several milk proteins: application of a new bioassay. J Dairy Sci 1998;81:909-17. ArticlePubMed
- 71. Shankaran M, Czerwieniec G, Fessler C, Wong PA, Killion S, Turner SM, et al. Dilution of oral D3-creatine to measure creatine pool size and estimate skeletal muscle mass: development of a correction algorithm. J Cachexia Sarcopenia Muscle 2018;9:540-6. ArticlePubMedPMCPDF
- 72. Stimpson SA, Turner SM, Clifton LG, Poole JC, Mohammed HA, Shearer TW, et al. Total-body creatine pool size and skeletal muscle mass determination by creatine-(methyl-D3) dilution in rats. J Appl Physiol (1985) 2012;112:1940-8. ArticlePubMed
- 73. Balsom PD, Söderlund K, Ekblom B. Creatine in humans with special reference to creatine supplementation. Sports Med 1994;18:268-80. ArticlePubMed
- 74. McCarthy C, Schoeller D, Brown JC, Gonzalez MC, Varanoske AN, Cataldi D, et al. D3-creatine dilution for skeletal muscle mass measurement: historical development and current status. J Cachexia Sarcopenia Muscle 2022 Sep 5 [Epub]. https://doi.org/10.1002/jcsm.13083. Article

 , Sanghee Park, Ph.D.2
, Sanghee Park, Ph.D.2 , Jiwoong Jang, Ph.D.1
, Jiwoong Jang, Ph.D.1 , Yeongmin Kim, M.S.3
, Yeongmin Kim, M.S.3 , Hee-Joo Kim, M.S.3
, Hee-Joo Kim, M.S.3







 E-submission
E-submission KSPEN
KSPEN KSSMN
KSSMN ASSMN
ASSMN JSSMN
JSSMN Cite
Cite